Publications
2026
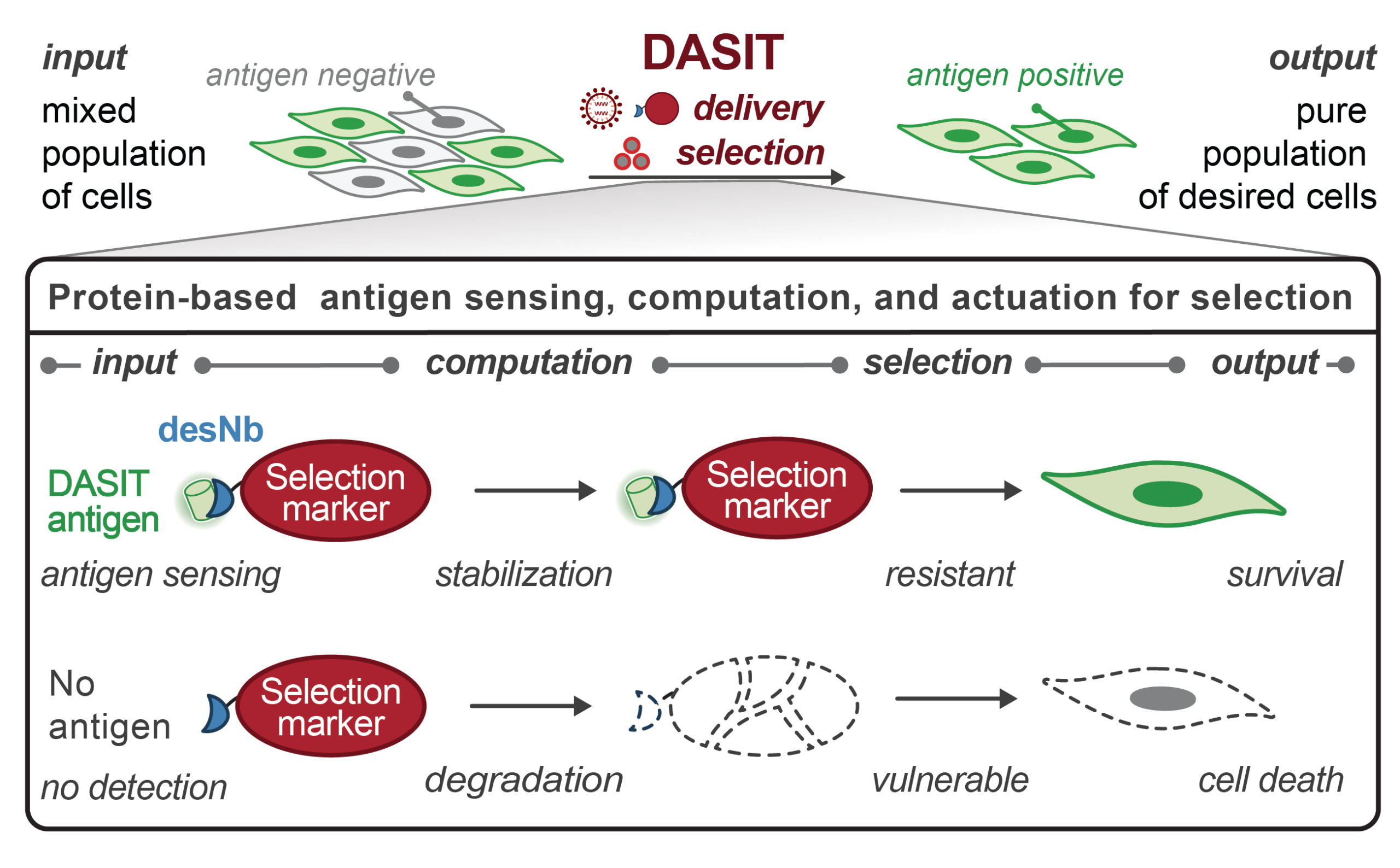
Programmable nanobody circuits for cell selection
Highlights
• DASIT enriches for antigen-positive cells across multiple selection markers and antigens
• Intermediate levels of DASIT expression support selection across stable and transient delivery modalities
• Logic-gated, precision genome engineering of human iPSCs via DASIT selection
• DASIT enables scalable activity-based selection for high-throughput base editing screens
•DASIT-selected engineered motor neurons survive grafting into acute spinal cord injury
2025
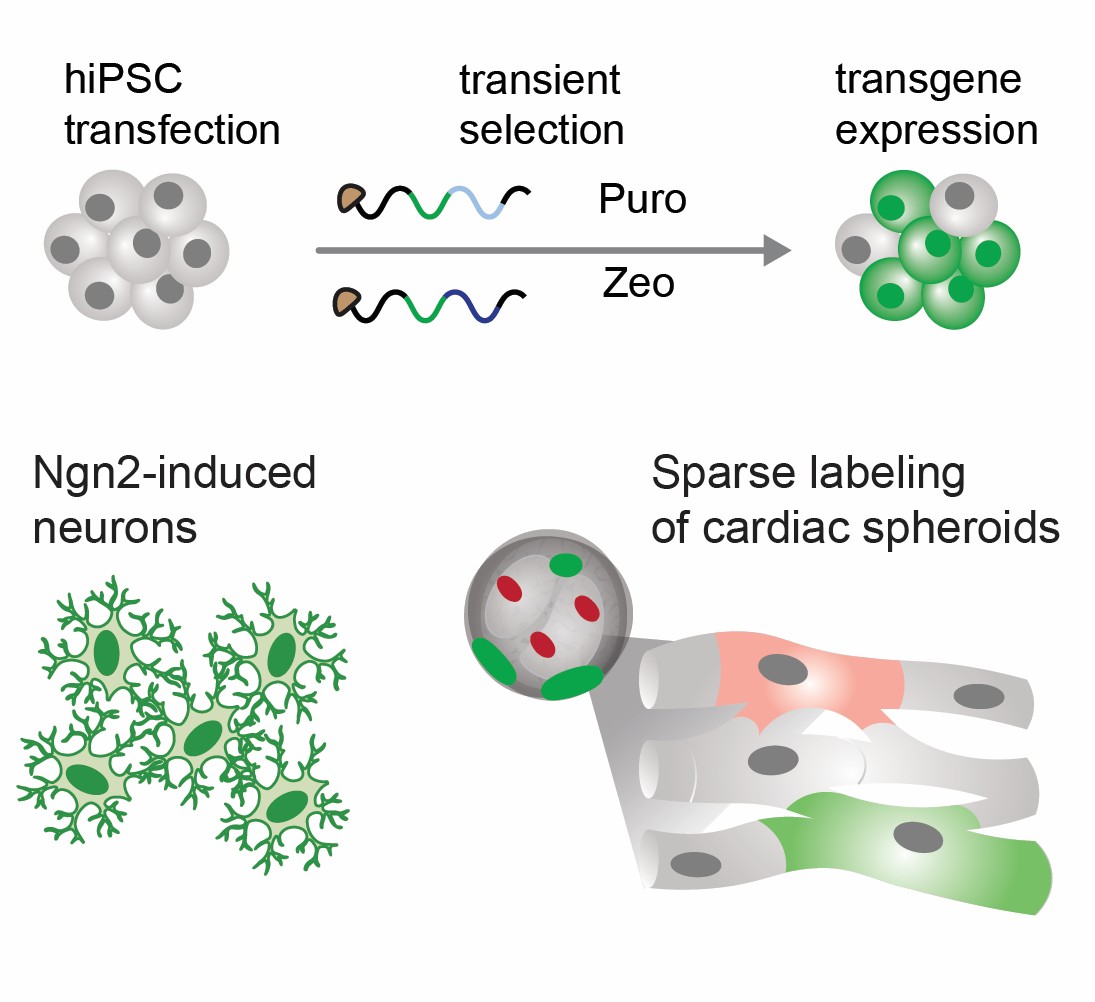
Self-amplifying RNA enables rapid, durable, integration-free programming of hiPSCs
Highlights
• A single saRNA transfection generates durable transgene expression
• saRNA transfection of Ngn2 in hiPSCs results in robust neuronal differentiation
• saRNA-delivery of functional reporters enables single-cell analysis of primary and hiPSC-derived cells
• saRNA-based sensor allows monitoring of maturation and drug responses in 3D cardiac spheroids
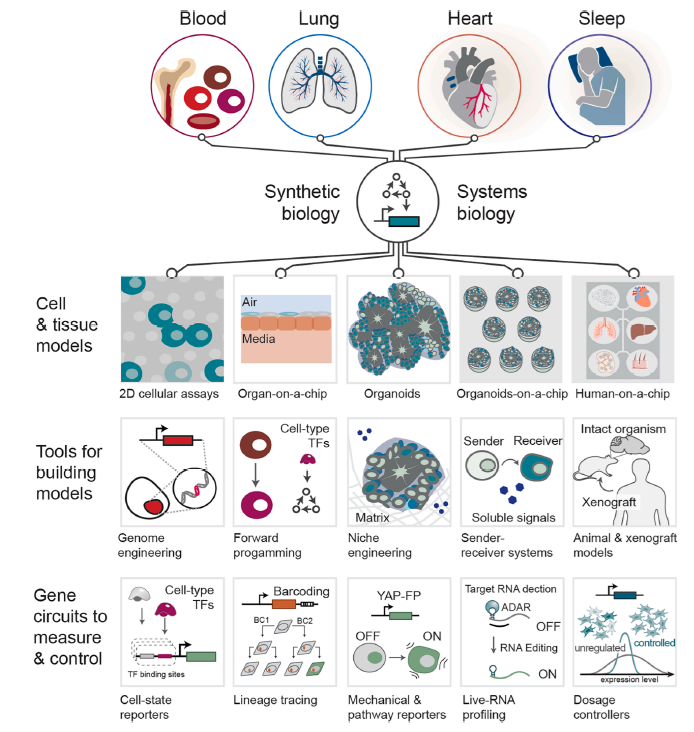
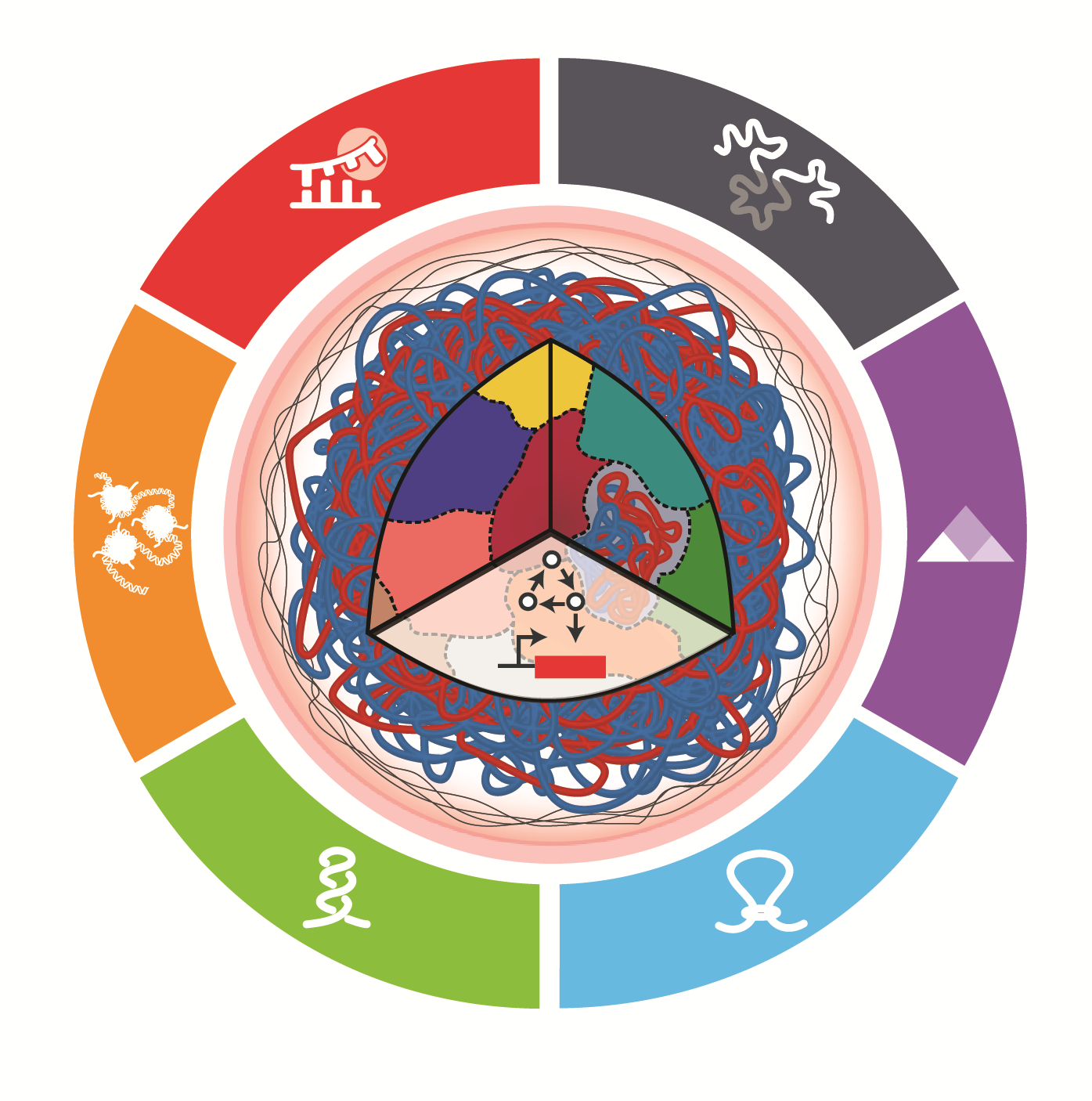
Integrating synthetic biology to understand and engineer the heart, lung, blood, and sleep systems
Chaikof EL, Chen J, Gillette MU, Boyer LA, Deans TL, Li P, Hilton IB, Daniels K, Goyal Y, Mei Y, Linghu C, Loveless TB, Truong DM, Blatchley MR, Gu M, Bashor CJ, Yang JH, Raman R, Reddy AB, Saha K, Davis J, Gupta K, Gao XJ*, Galloway KE*. * Co-corresponding
Cell Systems. 2025.
Synthetic biology offers control over cellular and tissue functions. As it moves beyond microbes into humans, synthetic biology enables precise control over gene expression, cell fate, and tissue organization across heart, lung, blood, and sleep systems. By integrating genome engineering, dynamic gene circuits, and high-dimensional biosensors, these advances support scalable, quantitative models of multicellular biology, expanding the need for systems-level models and integration. We highlight emerging systems such as tunable transcriptional regulators, synthetic organizers, and feedback circuits that bridge molecular control with functional outcomes. Furthermore, by combining omics data with artificial intelligence (AI)-guided circuit design, synthetic biology enables high-resolution cellular and tissue-scale models of development, cellular interactions, drug development, gene therapy, and therapeutic response. Key challenges remain—including delivery, transgene stability, and robust spatiotemporal control in physiologically relevant models. This perspective synthesizes field-spanning progress and defines shared priorities for engineering cells and tissues that function reliably across dynamic, multi-organ environments.
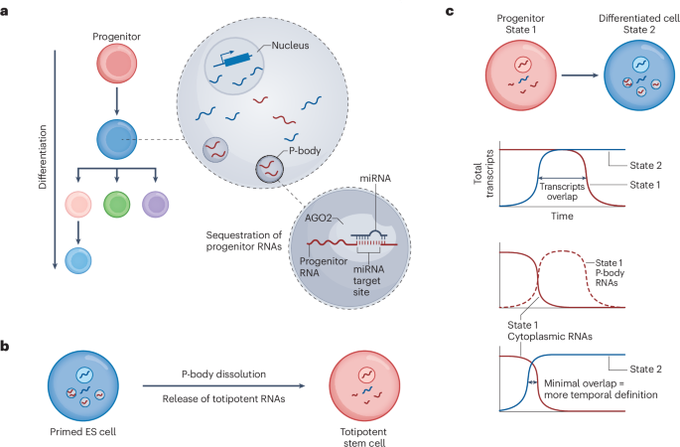
Modulating RNA condensates to control cell fate
Love KS and Galloway KE
Nature Biotechnology. 2025.
Highlight Pessina et al Nature Biotechnology. 2025.
RNA sequestration in P-bodies stabilizes cell-fate transitions by repressing past identities — a process that can be harnessed to control cell fate. The ability to control cellular identity is a central goal in biotechnology. In a paper in Nature Biotechnology, Pessina et al.1 investigate how developmental cell-fate transitions are influenced by RNA sequestration in cytoplasmic processing bodies (P-bodies). Comparing RNA profiles in the cytoplasm and in P-bodies for six cell types, including embryonic and differentiated states, the authors found that P-bodies are enriched for transcripts of the previous cell state across vertebrates. At a mechanistic level, cell-type-specific localization of RNA can be regulated by microRNA (miRNA). By manipulating RNA sequestration in P-bodies, the authors converted human embryonic stem cells into totipotent cells or germ cells. Future work could apply this localization strategy to enhance conversion to other cell states of interest or to improve temporal control of synthetic circuits.
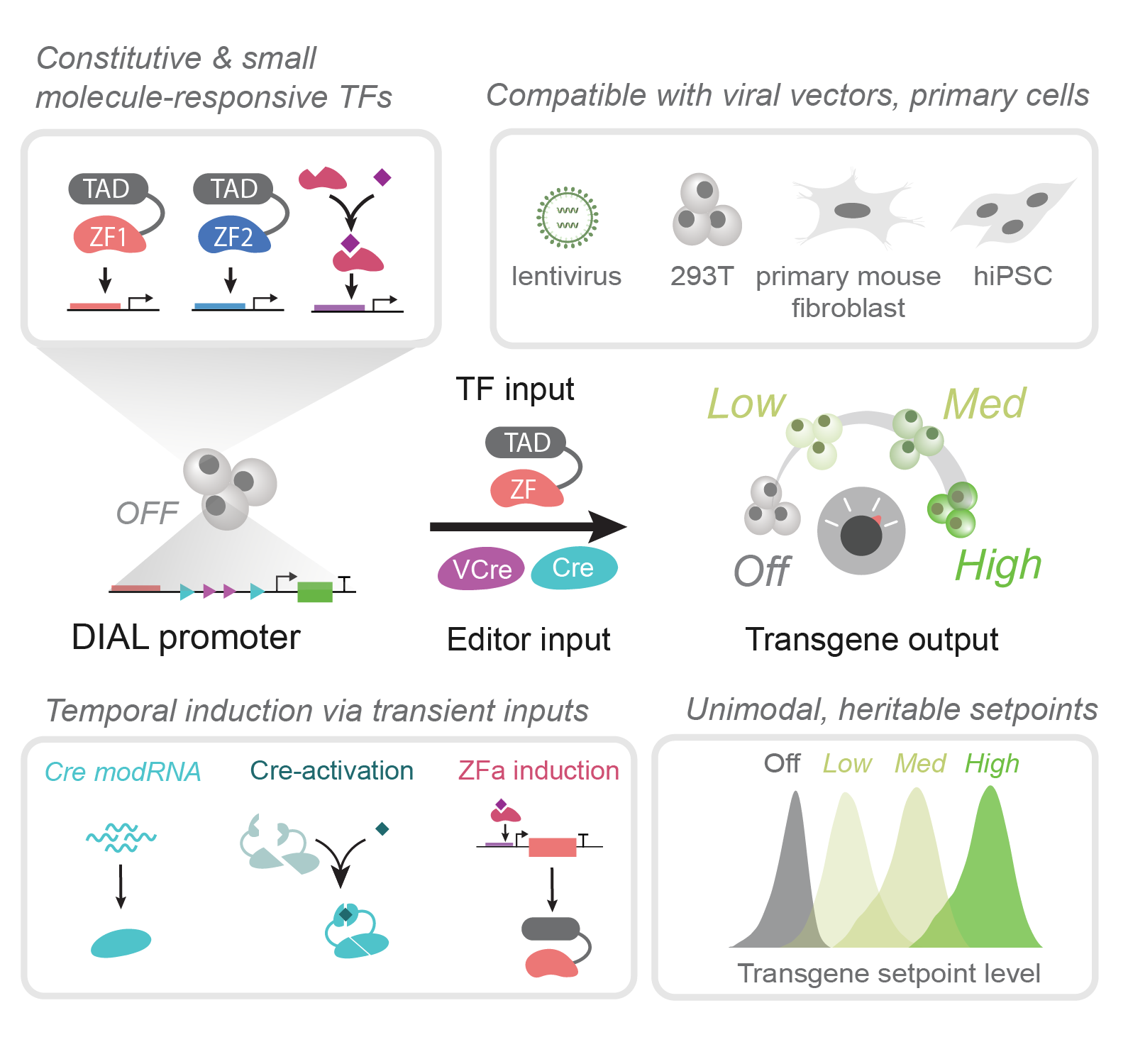
Programmable promoter editing for precise control of transgene expression
Kabaria SR, Bae Y, Ehmann ME, Lende-Dorn BA, Beitz AM, Peterman EL, Love KS, Ploessl DS, Galloway KE
Nature Biotechnology. 2025.
Highlights
- DIAL, a synthetic promoter system, edits the promoter to generate a range of unimodal setpoints
- DIAL setpoints are robust to varying levels of ZFa
- DIAL transmits transient inputs into heritable states
- TET-DIAL system enables small-molecule activation of defined setpoints
- DIAL controls expression in primary cells and iPSCs; regulates physiologically-relevant transgenes
- DIAL setpoints map fine-scale changes in regulator expression to rates of cell-fate transition
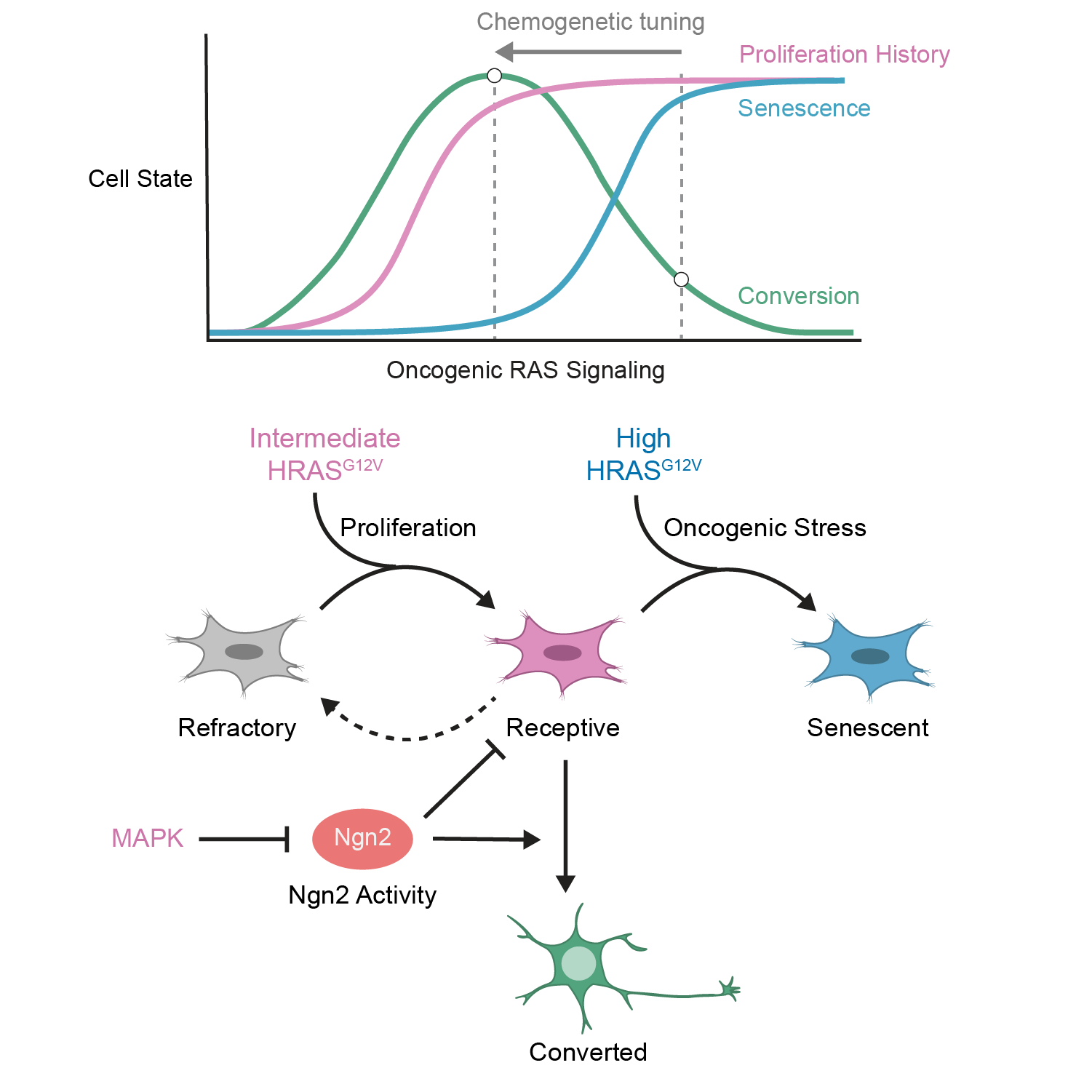
Chemogenetic tuning reveals optimal MAPK signaling for cell-fate programming
Lende-Dorn, BA, Atkinson, JC, Bae, Y and Galloway, KE
Cell Reports. 2025.
Highlights
- MAPK signaling drives proliferation and conversion of fibroblasts to motor neurons
- Cell-fate programming responds biphasically to HRASG12V expression
- A small-molecule MAPK inducer can replace HRASG12V for high rates of conversion
- MAPK signaling alters Ngn2 levels, influencing proliferation and conversion
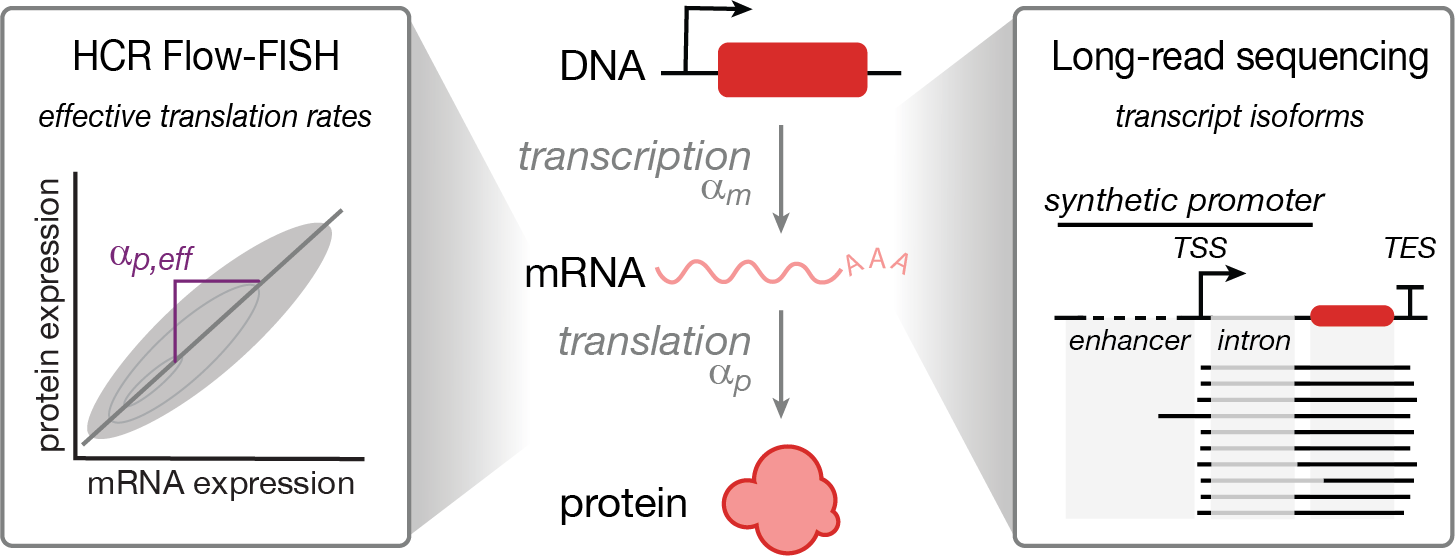
High-resolution profiling reveals coupled transcriptional and translational regulation of transgenes
Highlights
- HCR Flow-FISH enables simultaneous quantification of mRNA and protein levels in single cells
- Promoter sequences affect RNA transcript abundance and effective translation rate
- Choice of polyadenylation signal impacts effective translation rate of mRNA transcripts
- Gene coding sequence impacts effective translation rate but not mRNA levels
- Examining transcript isoforms highlights promoter-specific patterns of gene regulation
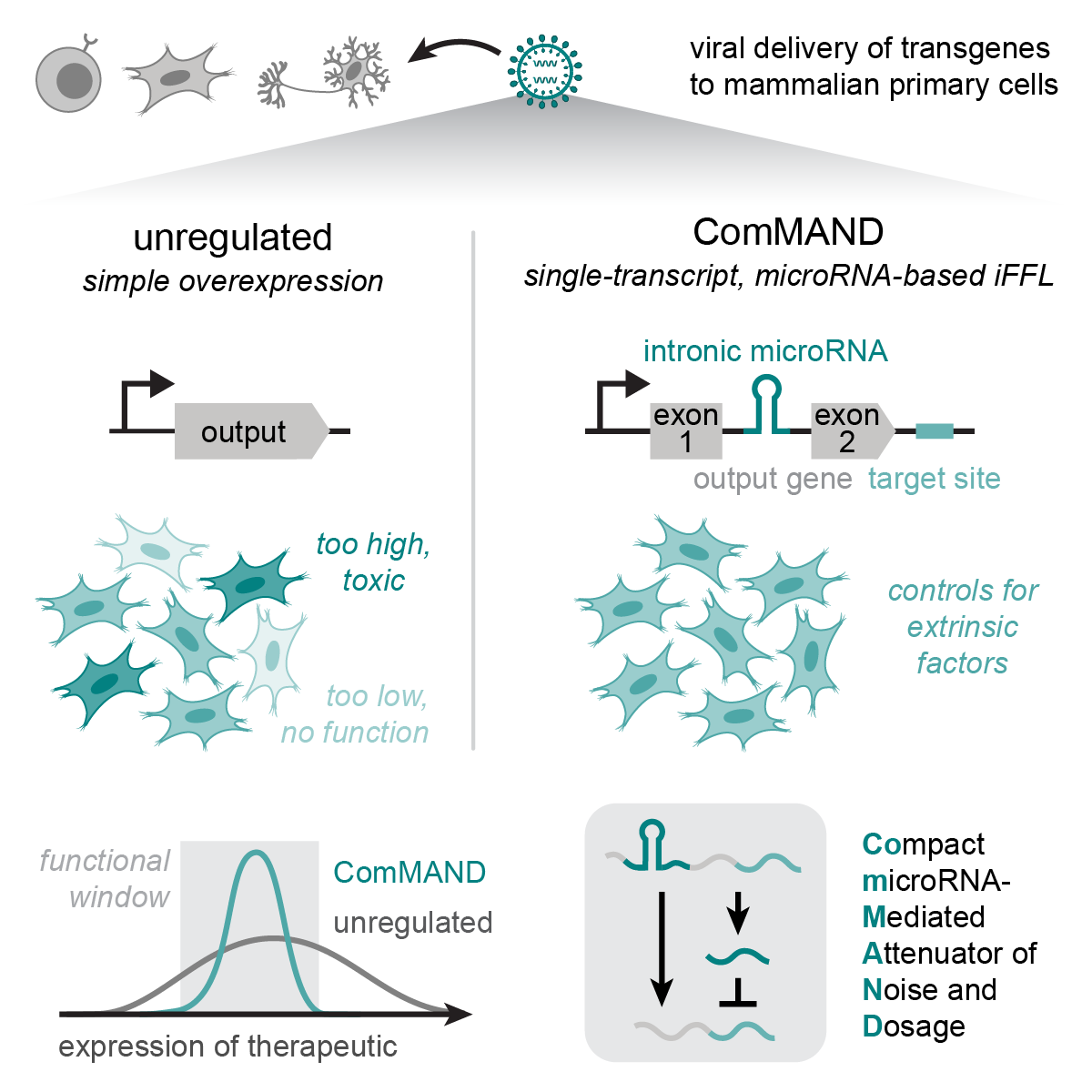
Model-guided design of microRNA-based gene circuits supports precise dosage of transgenic cargoes into diverse primary cells
Love KS, Johnstone CP, Peterman EL, Gaglione S, and Galloway KE
Cell Systems. 2025.
Highlights
- ComMAND precisely regulates output expression using a single-transcript, microRNA-based incoherent feedforward loop
- Selection of circuit components enables tuning of output expression
- A model of ComMAND activity identifies physiological limits and tuning strategies
- The single-transcript circuit matches or exceeds performance of two-gene implementations
- The single-transcript circuit regulates output expression across cell types and delivery methods
- ComMAND functions in diverse primary cells and can regulate clinically relevant output genes
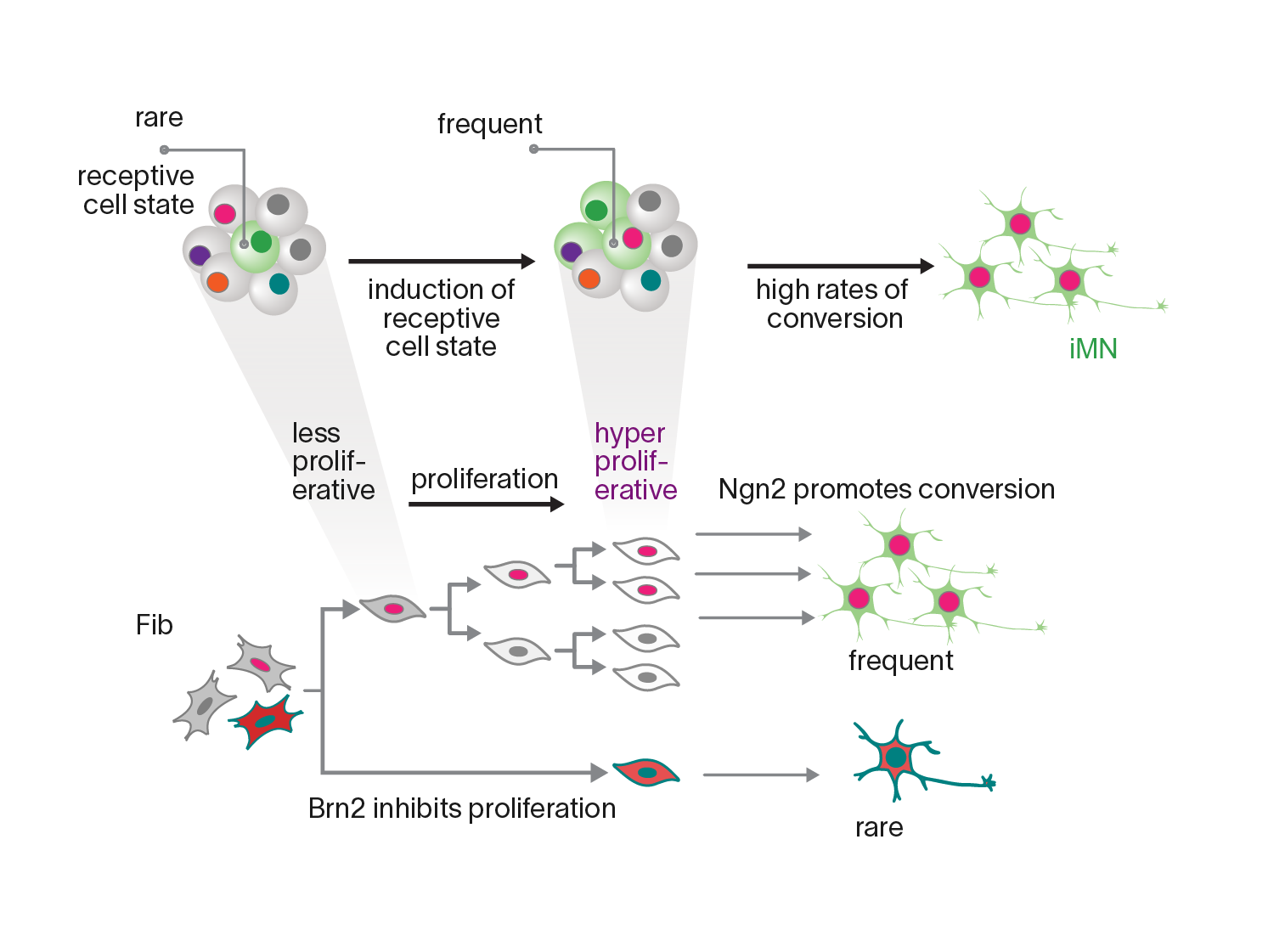
Proliferation history and transcription factor levels drive direct conversion to motor neurons
Highlights
- Proliferation history offers a principal axis to distinguish TF effect on fate
- Levels of individual TFs differentially influence the rate of conversion
- Proliferation history shapes the cell’s response to levels of pioneer TF, Ngn2
- Driving early hyperproliferation increases direct conversion of adult human fibroblasts
#synbio #gene_circuits #reprogramming #proliferation
Compact transcription factor cassettes generate functional, engraftable neurons by direct conversion
Wang NB, Adewumi HO, Lende-Dorn BA, Beitz AM, O’Shea TM, and Galloway, KE.
Cell Systems. 2025.
Highlights
- Compact, conversion cassettes are compatible with diverse delivery vectors
- Cocktail and conversion culture conditions influence the cell states of iMNs
- Optimized conversion cocktail supports neurotrophin-free conversion to iMN-like cells
- iMNs display electrical activity and graft in vivo within the central nervous system
#synbio #gene_circuits #reprogramming #proliferation
Engineered transcription factor binding arrays for DNA-based gene expression control in mammalian cells.
A Zouein, B Lende-Dorn, KE Galloway, T Ellis, F Ceroni
Trends in Biotechnology. 2024.
Highlights
Manipulating gene expression in mammalian cells is critical for cell engineering applications. Here we explore the potential of transcription factor (TF) recognition element arrays as DNA tools for modifying free TF levels in cells and thereby controlling gene expression. We first demonstrate proof-of-concept, showing that Tet TF-binding recognition element (RE) arrays of different lengths can tune gene expression and alter gene circuit performance in a predictable manner. We then open-up the approach to interface with any TF with a known binding site by developing a new method called Cloning Troublesome Repeats in Loops (CTRL) that can assemble plasmids with up to 256 repeats of any RE sequence. Transfection of RE array plasmids assembled by CTRL into mammalian cells show potential to modify host cell gene regulation at longer array sizes by sequestration of the TF of interest. RE array plasmids built using CTRL were demonstrated to target both synthetic and native mammalian TFs, illustrating the ability to use these tools to modulate genetic circuits and instruct cell fate. Together this work advances our ability to assemble repetitive DNA arrays and showcases the use of TF-binding RE arrays as a method for manipulating mammalian gene expression, thus expanding the possibilities for mammalian cell engineering.
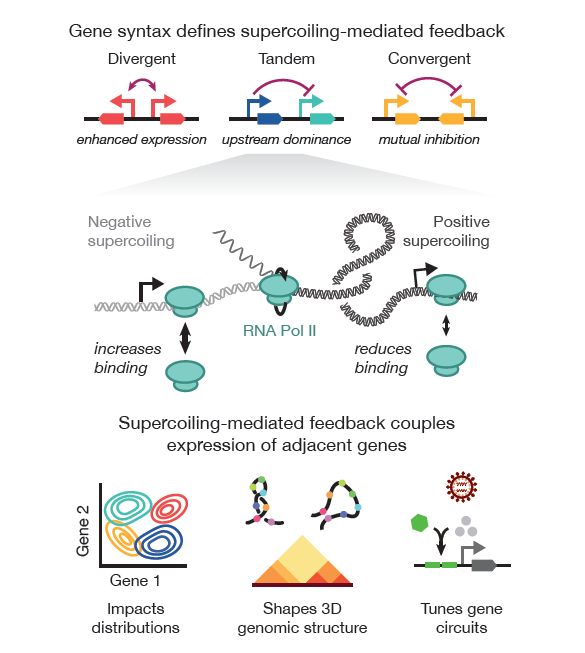
Gene syntax defines supercoiling-mediated transcriptional feedback
Highlights
- Transcription of an adjacent gene induces syntax-specific coupling
- Upstream dominance defines expression profiles of constitutive tandem transgenes
- Transcription induces syntax-specific structures across circuits and the surrounding locus
- Syntax-based tuning optimizes circuit expression and biologic production without part substitution
- Syntax augments performance of compact gene circuits across cell types
Tweetorial
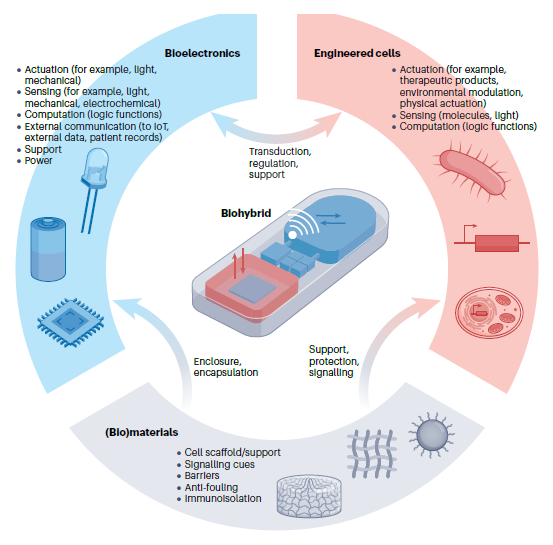
Integrating bioelectronics with cell-based synthetic biology
Abstract
Biohybrid devices based on engineered cells interfaced with bioelectronics represent a promising union where the strengths of each field can be synergistically combined, resulting in constructs with properties that are not otherwise achievable. Recent progress in biomaterials and cell-based synthetic biology has resulted in cells that can be remotely triggered via multiple modalities and can access a number of cellular pathways to achieve complex sensing and biomolecule production tasks. Although these living cells can be deployed as next-generation diagnostics and cell-based therapies, they are limited by the fundamental boundaries of biology. The parallel advances in synthetic biology, biomaterials and bioelectronics present new opportunities in devices for regulated cell therapies, diagnostic tools and next-generation robotics. In this Review, we discuss the enabling mechanisms of communication between engineered cells and bioelectronics platforms, describe the approaches and challenges in assembling and deploying such systems, and highlight recent prototypes. The continued advancement in cell support systems and both internal and external closed-loop control suggest forthcoming breakthrough opportunities for biohybrid bioelectronics.
Guaranteed multistability in a microRNA-based genetic network by formal methods
N. Nolan, E. Peterman, K. E. Galloway, R. M. Murray, E. D. Sontag, and D. Del Vecchio.
Proc. 2025 63rd IEEE Conference on Decision and Control (CDC). 2025.
The development of genetic memory devices in synthetic biology is a challenging process that requires extensive analysis and characterization. In mammalian systems, this complexity is compounded by the need for a small DNA payload for efficient delivery into the cell. Previous genetic memory devices have relied exclusively on protein-based regulation, which are limited by their large size; in this paper, we propose a microRNA-based multistable network, which effectively halves the payload size for more efficient delivery. We demonstrate that the system can be multistable, and use formal methods to characterize constraints on design parameters that guarantee multistability. Our results provide a new genetic network topology that can achieve multistability and demonstrate the use of formal methods in the design of sophisticated genetic network architectures against non-convex top-level specifications.
2024
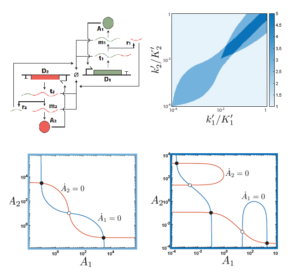
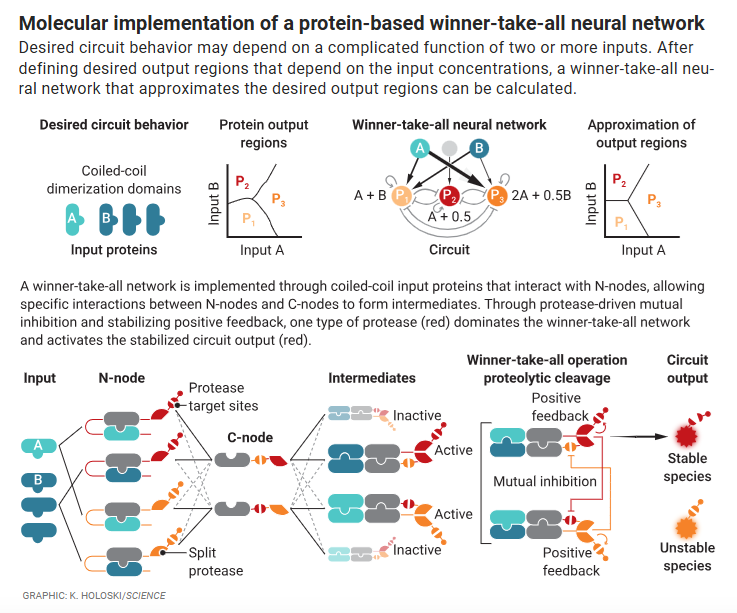
Bringing neural networks to life
Galloway KE and Johnstone CJ
Science. 2024.
Highlight Chen et al Science. 2024.
Native gene circuits that drive differentiation and maintain cell fate are constructed from layers of multi-input programs that compute cell responses to diverse stimuli. Although multilayer synthetic circuits have been constructed with DNA oligonucleotides in vitro , in bacteria, and in mammalian cells, adaptation of these methods to new tasks often requires reselection and reoptimization of circuit components. Chen et al. describe a protein-level synthetic circuit framework that implements a “winner-take-all” neural network capable of classifying the relative abundance of multiple inputs to modify the circuit output. This network replicates various classification circuits by adjusting the relative concentrations of a few components. By connecting this circuit to molecular pathways that regulate apoptosis, the authors demonstrate the promise of this approach for programming cell fate outcomes. This could be extended to design complex neural circuits that augment the computational capacity of cells.
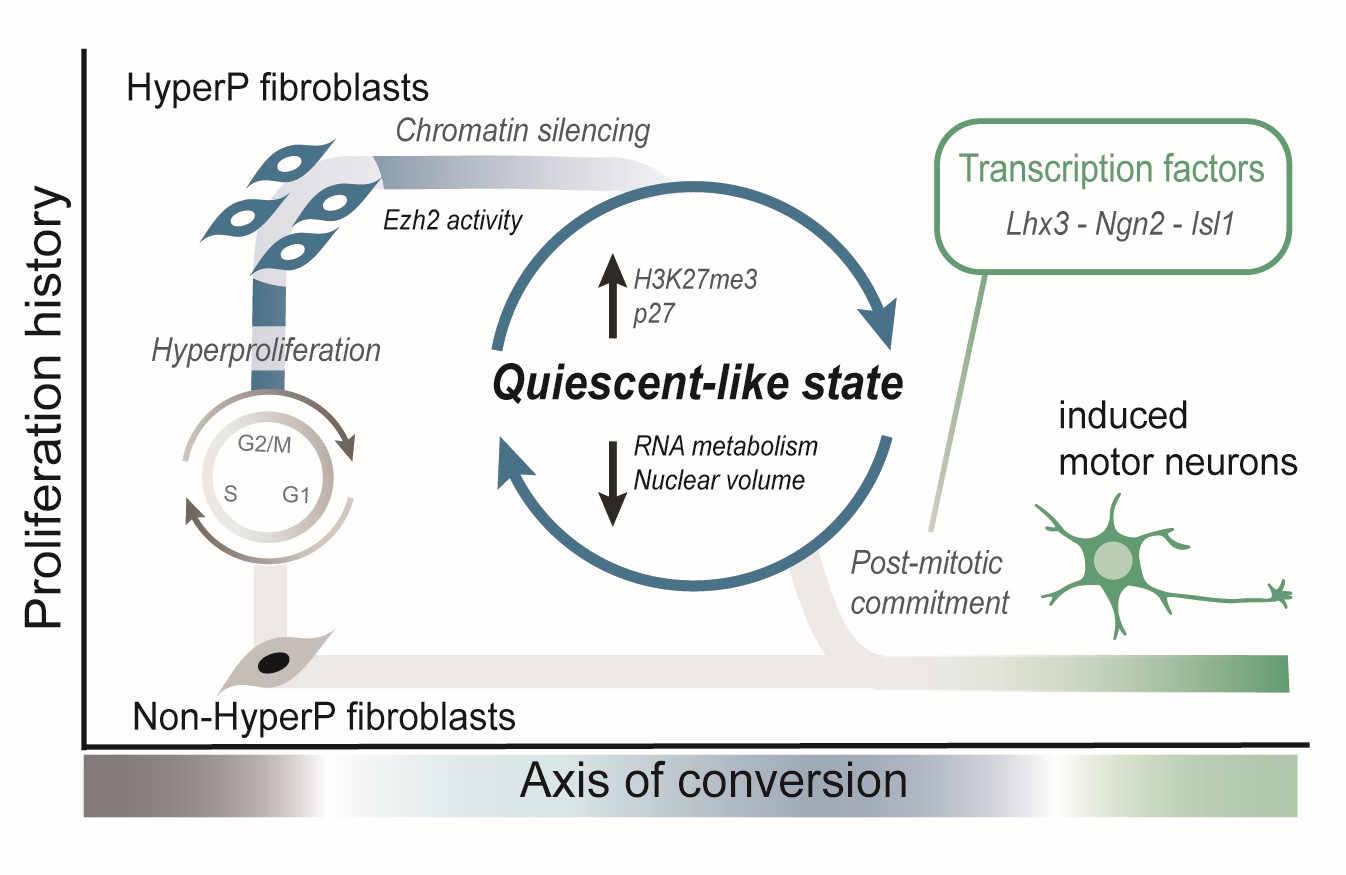
Cells transit through a quiescent-like state to convert to neurons at high rates
Beitz AM, Teves, J, Oakes, CG, Johnstone, CP Wang, NB, Brickman,JM and Galloway, KE
bioRxiv. 2024.
Highlights
- Proliferation drives cells to a compact nuclear state that is receptive to TF-mediated conversion.
- Increased receptivity to TFs corresponds to reduced nuclear volumes.
- Reprogrammable cells display global, genome-wide increases in H3K27me3.
- High levels of H3K27me3 support cells’ transits through a state of altered RNA metabolism.
- Inhibition of Ezh2 increases nuclear size, reduces the expression of the quiescence marker p27.
- Acute inhibition of Ezh2 abolishes motor neuron conversion.
Tweetorial
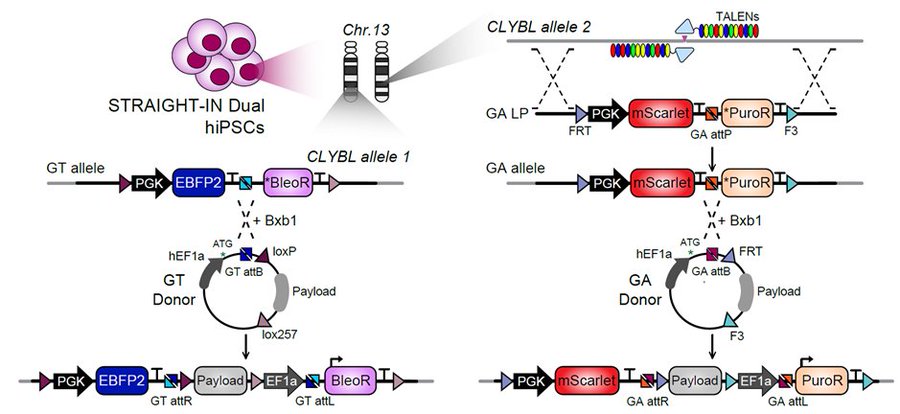
STRAIGHT-IN Dual: a platform for dual, single-copy integrations of DNA payloads and gene circuits into human induced pluripotent stem cells
Blanch Ascensio, A, Ploessl DS, Wang, NB, Mummery, CL, Galloway KE* and Davis, RP
bioRxiv. 2024.
Highlights
- STRAIGHT-IN Dual enables simultaneous, allele-specific, single-copy integration of two DNA payloads
- STRAIGHT-IN Dual generates uniformly integrated cells (100% efficiency) within one week
- STRAIGHT-IN Dual allowed us to identify and overcome sources of transgene silencing in hiPSCs
- We demonstrate the optimization of a system for inducible forward programming to induced neurons
- We demonstrate control of two functionally-relevant transgenes via orthogonal inducible systems
Changes in cell-cycle rate drive diverging cell fates
Galloway, KE
Nature Reviews Genetics. 2024.
Highlight Kueh et al Science. 2013.
Changes in the concentrations of transcription factors drive cells to commit to different fates. These concentration changes can be achieved through mechanisms such as transcriptional activation or protein degradation. However, the role of the cell cycle in regulating transcription factor levels and cell fate remains underappreciated. During differentiation, cells often divide before committing to divergent fates, enabling the generation of new cells and maintenance of a self-renewing progenitor population. Differences in the proliferation rate of these populations emerge as their fates diverge. Thus, it remains challenging to separate the role of the cell cycle in establishing cell identity from cell identity itself. In 2013, to address how cell cycle influences the divergence of myeloid and lymphoid cells from a progenitor population, Kueh et al. examined how expression of the pioneer transcription factor PU.1 generated distinct cell fates in mice.
Rewinding the tape to identify intrinsic determinants of reprogramming potential
Galloway, KE
Cellular Reprogramming. 2024.
Highlight Jain et al Cell Systems. 2024.
Via retrospective isolation of clones using Rewind, Jain et al. identified primed states of cells that reprogram to induced pluripotent stem cells. Examining clones, they find that cells retain memory of over several rounds of cell division. Moreover, they show that extrinsic factors change the number of primed cells, suggesting that there exist diverse paths of reprogramming and states of priming.
Guaranteeing system-level properties in genetic circuits subject to context effects
Highlights
The identification of constraints on system parameters that will ensure that a system achieves desired requirements remains a challenge in synthetic biology, where components unintentionally affect one another by perturbing the cellular environment in which they operate. This paper shows how to solve this problem optimally for a class of input/output system-level specifications, and for unintended interactions due to resource sharing. Specifically, we show how to solve the problem based on the input/output properties of the subsystems and on the unintended interaction map. Our approach is based on the elimination of quantifiers in monotone properties of the system. We illustrate applications of this methodology to guaranteeing system-level performance of multiplexed and sequential biosensing and of bistable genetic circuits.
Accelerating Diverse Cell-Based Therapies Through Scalable Design
Highlights
- Advances in synthetic biology, genomics, and automation are needed to translate transgenic systems to primary cells and stem cells at clinically relevant scales.
- Techniques optimized for biologic production are not well-suited for cell-based therapies.
- Offline design and engineering of allogeneic cells will improve the production speed and scalability of stem cell–based therapies.
- Co-design of genetic cargo and transgenesis methods can improve engineering for a given cell type.
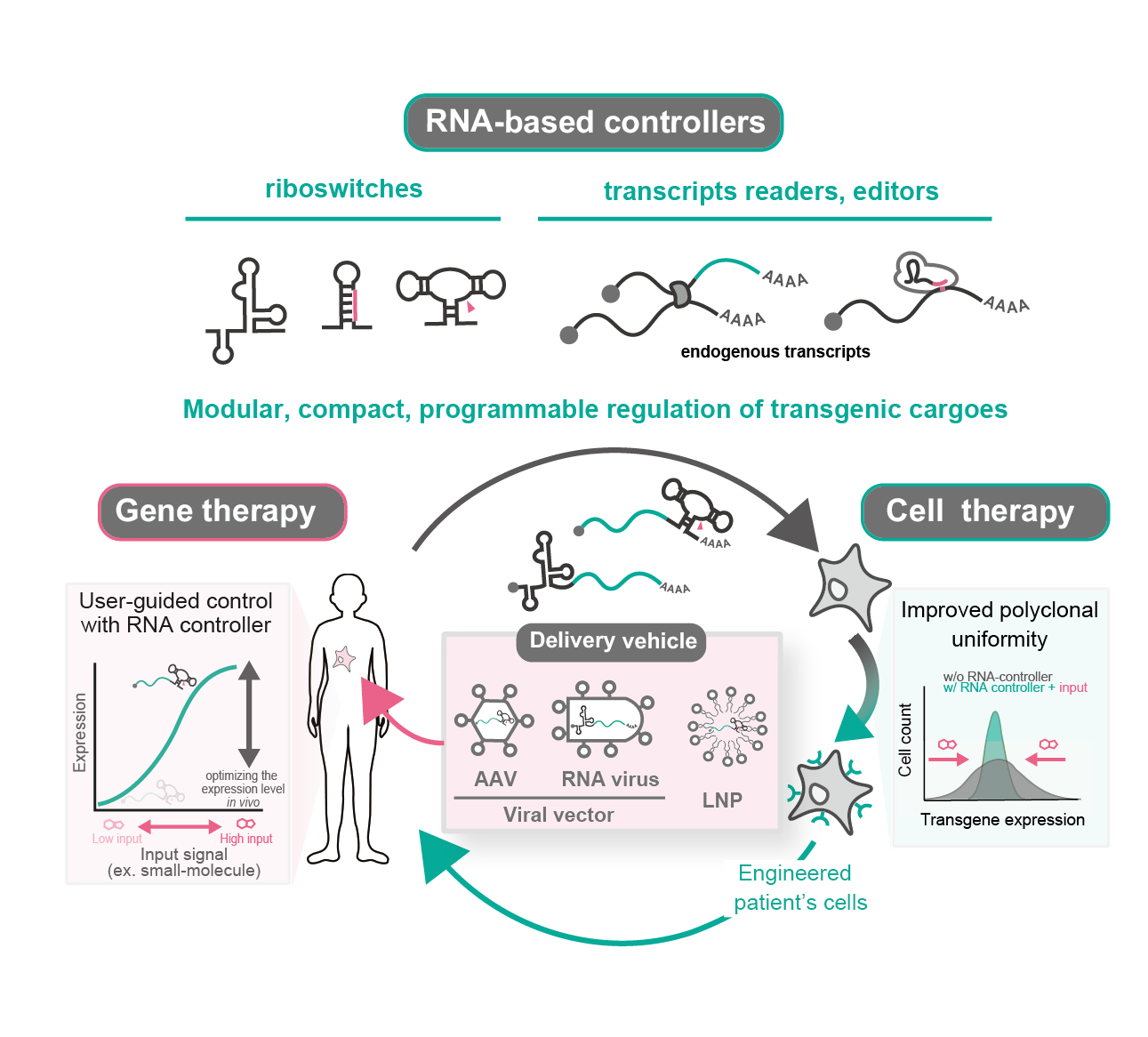
RNA-based controllers for engineering gene and cell therapies
Current Opinion in Biotechnology. 2024.
Highlights
- RNA offers highly compact, modular, portable, and programmable gene regulation.
- Ligand-responsive riboswitches allow for user-guided control of transgenes.
- RNA-responsive RNA controllers detect and respond to transcriptional state.
#synbio #gene_circuits #RNA #cell_therapies
2022
The sound of silence: Transgene silencing in mammalian cell engineering
Cabera, A* , Edelstein, HI*, Glykofrydis, F*, Love, KS*, Palacios, S* Tycko, J*, Zhang, M*, Lensch, S, Shields, CE, Livingston, M, Weiss, R, Zhao, H, Haynes, KA, Morsut, L, Chen, YY, Khalil, AS, Wong, WW, Collins, JJ, Rosser, SJ, Karen Polizzi, K, Elowitz, MB, Fussenegger, M, Hilton, IB, Leonard, JN, Bintu, L, Galloway, KE, Deans, TL.
Cell Systems. 2022.
Summary
To elucidate principles operating in native biological systems and to develop novel biotechnologies, synthetic biology aims to build and integrate synthetic gene circuits within native transcriptional networks. The utility of synthetic gene circuits for cell engineering relies on the ability to control the expression of all constituent transgene components. Transgene silencing, defined as the loss of expression over time, persists as an obstacle for engineering primary cells and stem cells with transgenic cargos. In this review, we highlight the challenge that transgene silencing poses to the robust engineering of mammalian cells, outline potential molecular mechanisms of silencing, and present approaches for preventing transgene silencing. We conclude with a perspective identifying future research directions for improving the performance of synthetic gene circuits.
Supercoiling-mediated feedback rapidly couples and tunes transcription
Johnstone, CP and Galloway, KE.
Cell Reports. 2022.
Highlights
- Supercoiling dynamics confer rapid, tunable coupling between adjacent genes
- Syntax—the relative order and orientation of genes—alters expression levels
- Supercoiling-dependent feedback tunes transcriptional variance and bursting
- Supercoiling coordinates the dynamics of transcriptional networks and gene circuits
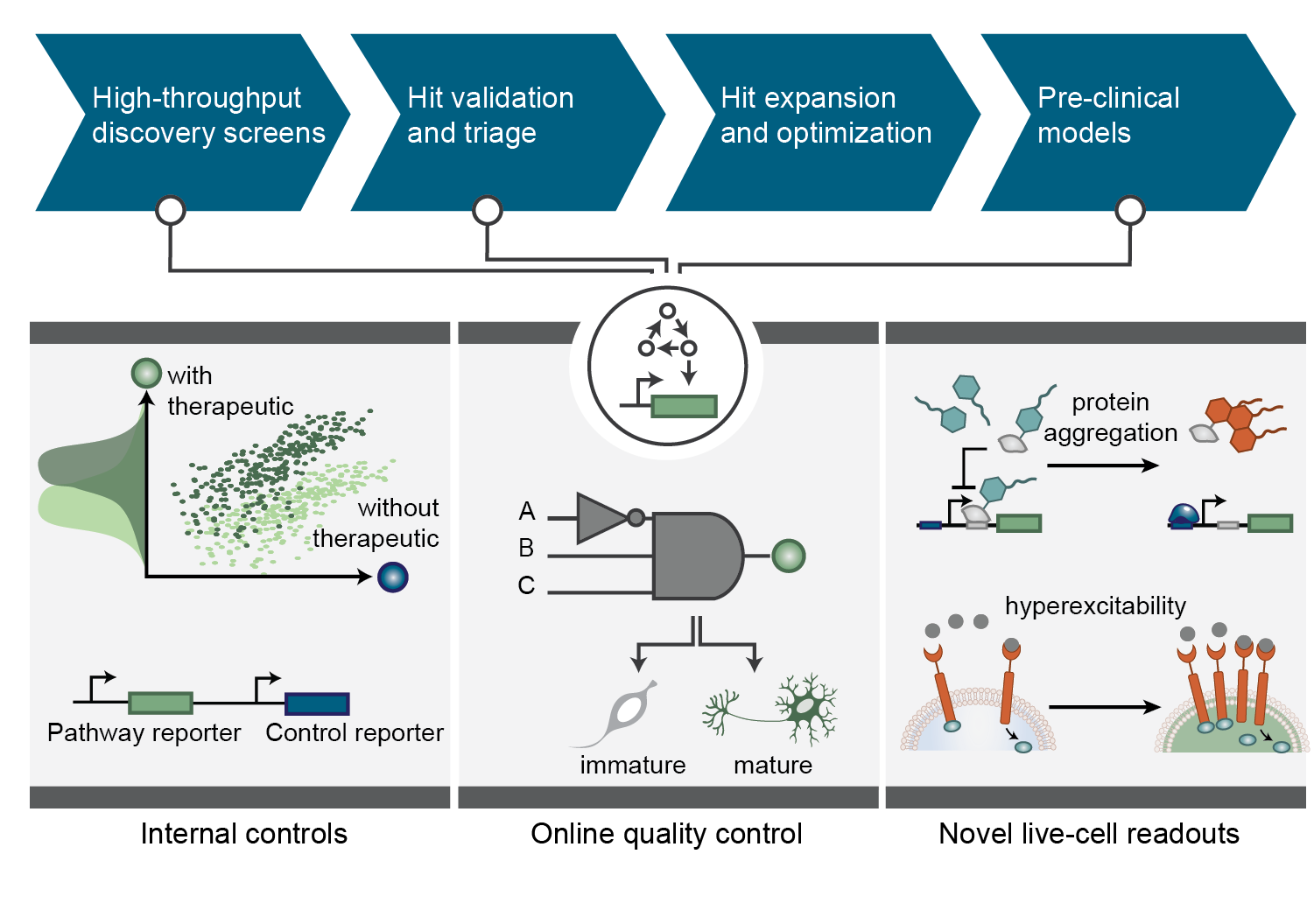
Synthetic gene circuits as tools for drug discovery
Beitz, AM, Oakes, CG and Galloway, KE.
Trends in Biotechnology. 2022.
Highlights
- Synthetic gene circuits enable real-time monitoring of diverse molecular events in live cells.
- Synthetic circuits provide internal controls to expedite hit validation and facilitate hit prioritization.
- Synthetic circuits enable on-line validation of induced pluripotent stem cell (iPSC)-derived cells.
- Synthetic circuits potentiate longitudinal tracking of neurodegenerative-associated phenotypes.
2021
Engineering cellular symphonies out of transcriptional noise
Johnstone, CP and Galloway, KE.
Nature Reviews Molecular Cell Biology. 2021.
Summary
Development unfolds through a series of orchestrated spatial and temporal gene-expression patterns. Despite relying on the noisy process of transcription, expression patterns remain robust to myriad disturbances. To achieve the goal of building complex tissues from the bottom up, synthetic biology must learn how to buffer and harness transcriptional noise.
2020
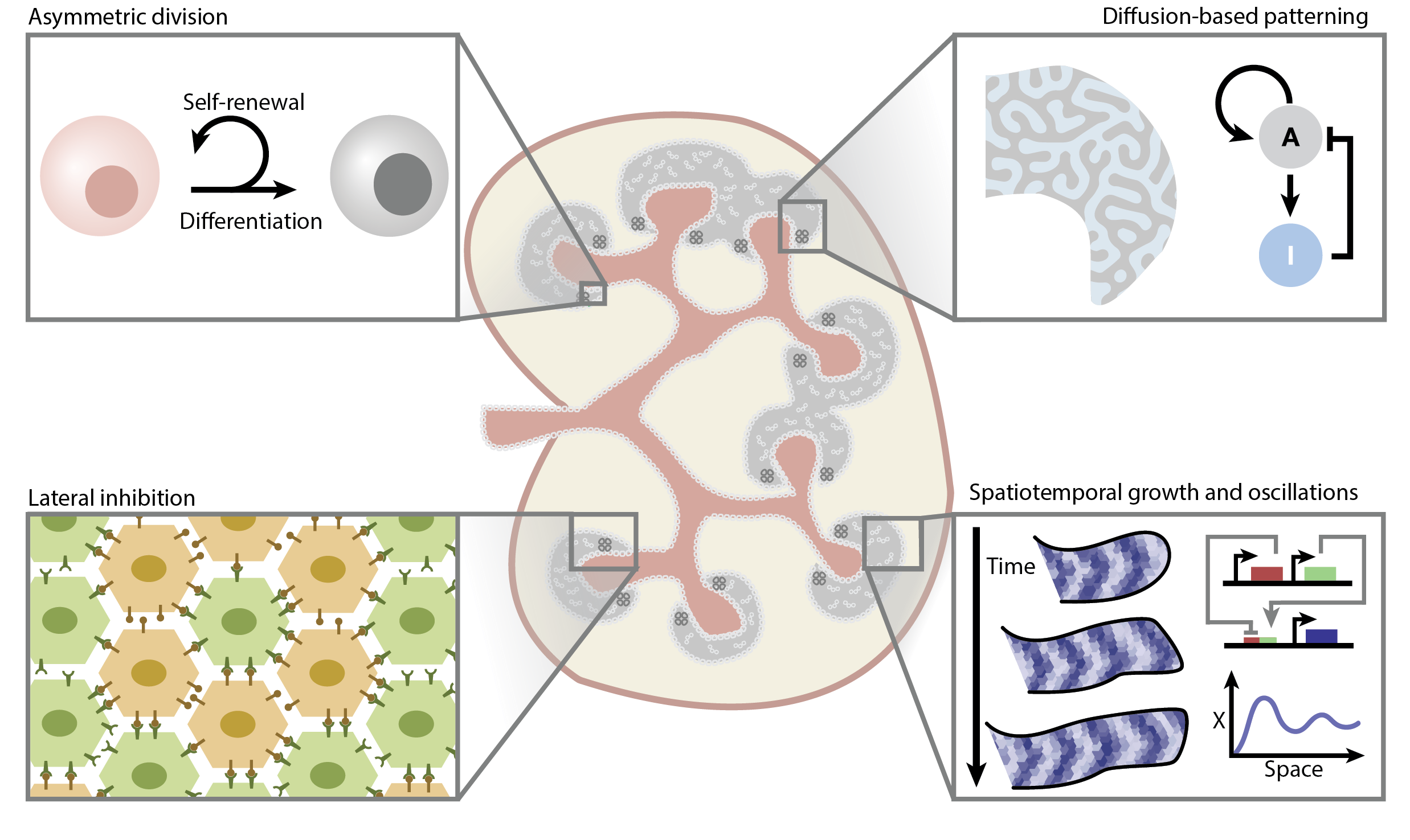
Understanding and engineering chromatin as a dynamical system across length and time scales.
Johnstone, CP*, Wang, NB*, Sevier, SA, and Galloway, KE.
Cell Systems. 2020.
Summary
Connecting the molecular structure and function of chromatin across length and timescales remains a grand challenge to understanding and engineering cellular behaviors. Across five orders of magnitude, dynamic processes constantly reshape chromatin structures, driving spaciotemporal patterns of gene expression and cell fate. Through the interplay of structure and function, the genome operates as a highly dynamic feedback control system. Recent experimental techniques have provided increasingly detailed data that revise and augment the relatively static, hierarchical view of genomic architecture with an understanding of how dynamic processes drive organization. Here, we review how novel technologies from sequencing, imaging, and synthetic biology refine our understanding of chromatin structure and function and enable chromatin engineering. Finally, we discuss opportunities to use these tools to enhance understanding of the dynamic interrelationship of chromatin structure and function.
Engineering cell fate: Applying synthetic biology to cellular reprogramming.
Wang, NB, Beitz, AM, Galloway, KE.
Current Opinion in Systems Biology. 2020.
Highlights
- Roadblocks to cellular reprogramming can be overcome with synthetic biology tools.
- Highly interconnected aspects of latent donor cell identity affect reprogramming.
- Recent systems-level studies of reprogramming identify key drivers of reprogramming.
- Advances in synthetic biology offer new tools for coordinating reprogramming processes.
Prior to 2020
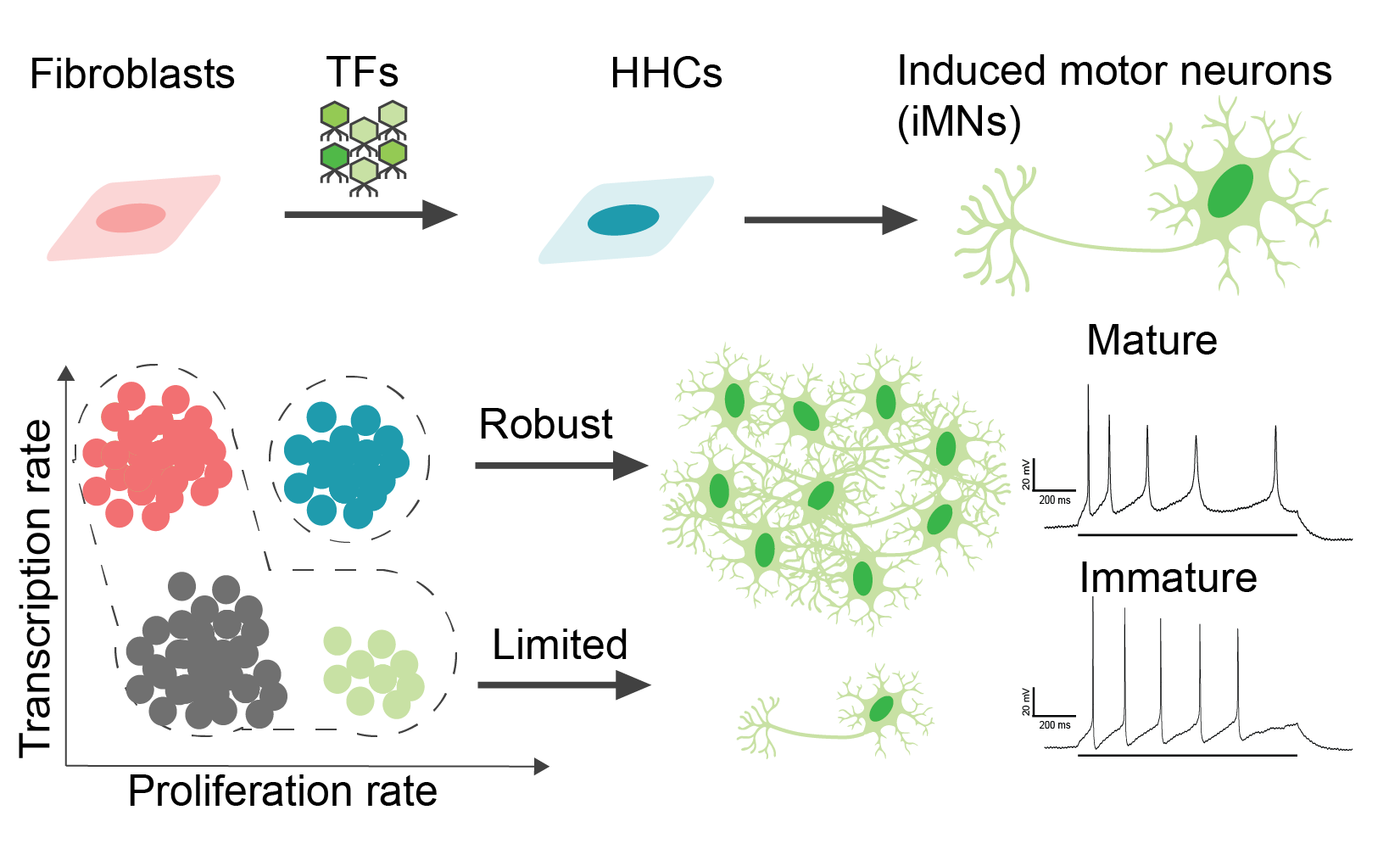
Mitigating antagonism between transcription and proliferation mediated near-deterministic reprogramming
Highlights
- Chemical, genetic cocktail reduces genomic stress induced by TF reprogramming
- DDRR cocktail expands population of hypertranscribing, hyperproliferating cells (HHCs)
- Supported by topoisomerases, HHCs reprogram at near-deterministic rates
- Topoisomerase expression reduces negative DNA supercoiling and R-loop formation
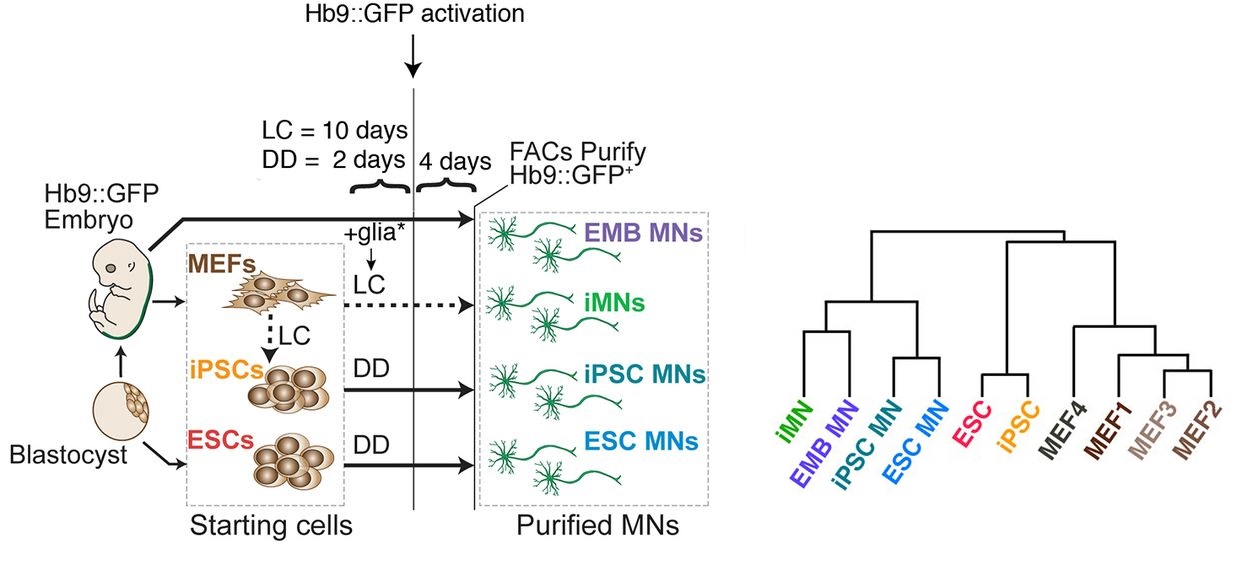
Comparative genomic analysis of embryonic, lineage-converted, and stem cell-derived motor neurons.
Abstract
Advances in stem cell science allow the production of different cell types in vitro either through the recapitulation of developmental processes, often termed ‘directed differentiation’, or the forced expression of lineage-specific transcription factors. Although cells produced by both approaches are increasingly used in translational applications, their quantitative similarity to their primary counterparts remains largely unresolved. To investigate the similarity between in vitro-derived and primary cell types, we harvested and purified mouse spinal motor neurons and compared them with motor neurons produced by transcription factor-mediated lineage conversion of fibroblasts or directed differentiation of pluripotent stem cells. To enable unbiased analysis of these motor neuron types and their cells of origin, we then subjected them to whole transcriptome and DNA methylome analysis by RNA sequencing (RNA-seq) and reduced representation bisulfite sequencing (RRBS). Despite major differences in methodology, lineage conversion and directed differentiation both produce cells that closely approximate the primary motor neuron state. However, we identify differences in Fas signaling, the Hox code and synaptic gene expression between lineage-converted and directed differentiation motor neurons that affect their utility in translational studies.
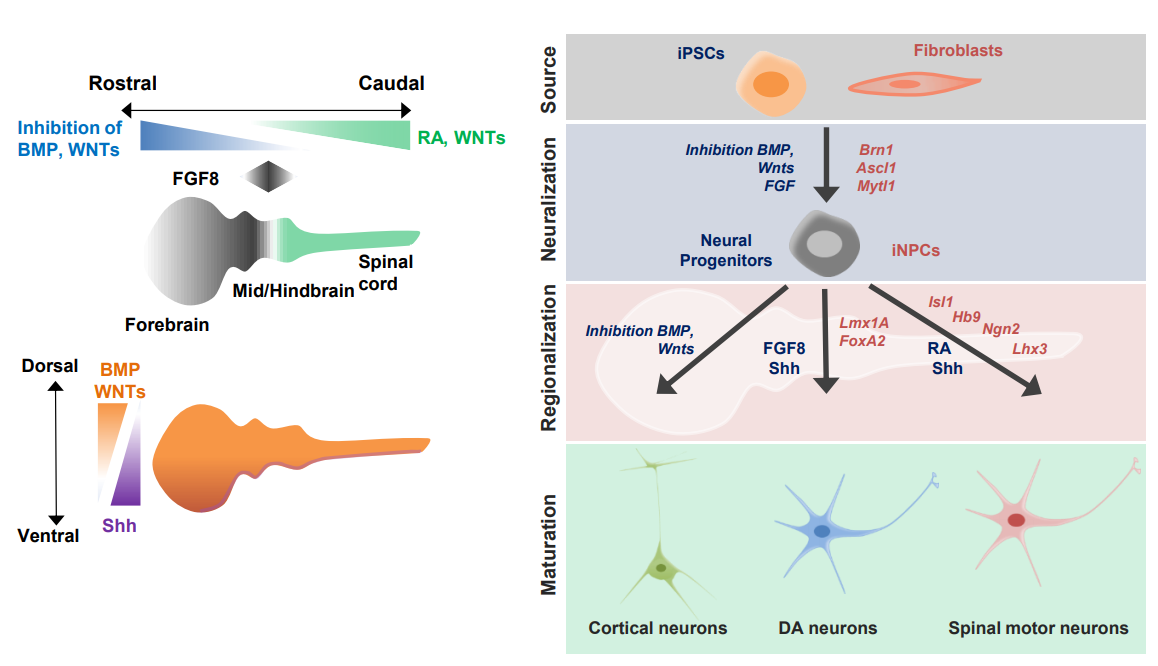
Modeling neurodegenerative diseases and neurodevelopmental disorders with reprogrammed cells
Summary
Induced pluripotent stem cells and reprogrammed cells offer the opportunity to generate disease-relevant human cells from readily-available patient cells for studying disease and identifying therapeutics.
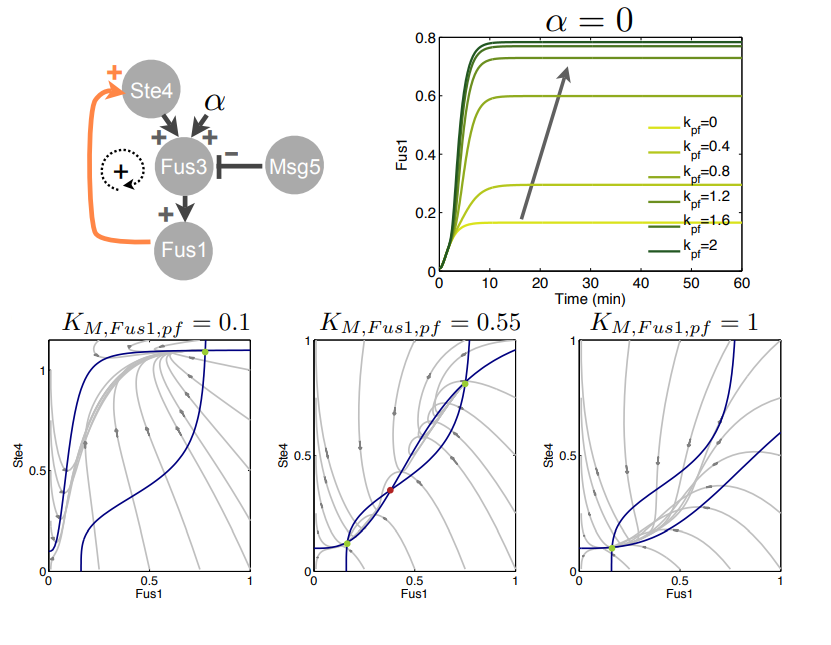
Feedback loops in biological networks
Abstract
We introduce fundamental concepts for the design of dynamics and feedback in molecular networks modeled with ordinary differential equations. We use several examples, focusing in particular on the mitogen-activated protein kinase (MAPK) pathway, to illustrate the concept that feedback loops are fundamental in determining the overall dynamic behavior of a system. Often, these loops have a structural function and unequivocally define the system behavior. We conclude with numerical simulations highlighting the potential for bistability and oscillations of the MAPK pathway re-engineered through synthetic promoters and RNA transducers to include positive and negative feedback loops.
Download
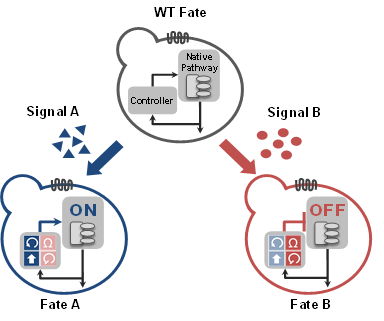
Dynamically reshaping signaling networks to program cell fate via genetic controllers
Galloway, KE, Franco, E, and Smolke, CD.
Science. 2013.
Abstract
Engineering of cell fate through synthetic gene circuits requires methods to precisely implement control around native decision-making pathways and offers the potential to direct cell processes. We demonstrate a class of genetic control systems, molecular network diverters, that interface with a native signaling pathway to route cells to divergent fates in response to environmental signals without modification of native genetic material. A method for identifying control points within natural networks is described that enables the construction of synthetic control systems that activate or attenuate native pathways to direct cell fate. We integrate opposing genetic programs by developing network architectures for reduced antagonism and demonstrate rational tuning of performance. Extension of these control strategies to mammalian systems should facilitate the engineering of complex cellular signaling systems.
Synthetic biology: Advancing biological frontiers by building synthetic systems.
Chen, YY*, Galloway, KE*, and Smolke, CD.
Genome Biology. 2012.
Abstract
Synthetic biology is an emerging field of interdisciplinary research that seeks to transform our ability to probe, manipulate, and interface with living systems by combining the knowledge and techniques of biology, chemistry, computer science, and engineering. Its main aim is to increase the ease and efficiency with which biological systems can be designed, constructed, and characterized. Core efforts in the field have focused on the development of tools to support this goal, including new approaches to biological design and fabrication. Although the first generation of synthetic systems demonstrated genetic circuits that encode dynamic behavior, cellular computational operations, and biological communication channels, more recent research has focused on implementing synthetic biological devices and systems in diverse applications, including disease therapy, environmental remediation, and biosynthesis of commodity chemicals. As the field matures, synthetic biology is advancing biological frontiers by expanding biomanufacturing capabilities, developing next-generation therapeutic approaches, and providing new insights into natural biological systems. Here, we review the theoretical foundations, diverse tool kits, and engineered systems that have emerged from synthetic biology and discuss current as well as potential future applications, which include in-depth studies of basic biology (such as understanding endogenous signaling pathways and feedback circuits) and new frontiers in health and medicine (such as identification of diseased cells and targeted therapeutics).
Temperature Responsive Biopolymer for Mercury Remediation.
Kostal, J, Mulchandani, A, Gropp, KE, and Chen, WA.
Environmental Science & Technology. 2003.
Abstract
Tunable biopolymers based on elastin-like polypeptides (ELP) were engineered for the selective removal of mercury. ELP undergoes a reversible thermal precipitation within a wide range of temperatures and was exploited to enable easy recovery of the sequestered mercury. A bacterial metalloregulatory protein, MerR, which binds mercury with an unusually high affinity and selectivity, was fused to the ELP to provide the highly selective nature of the biopolymers. Selective binding of mercury was demonstrated at an expected ratio of 0.5 mercury/biopolymer, and minimal binding of competing heavy metals (cadmium, nickel, and zinc), even at 100-fold excess, was observed. The sequestered mercury was extracted easily, enabling continuous reuse of the biopolymers. In repeating cycles, mercury concentration was reduced to ppb levels, satisfying even drinking water limits. Utility of the biopolymers with mercury-contaminated Lake Elsinore water was demonstrated with no decrease in efficiency. The nanoscale biopolymers reported here using metalloregulatory proteins represent a “green” technology for environmentally benign mercury removal. As nature offers a wide selection of specific metalloregulatory proteins, this technology offers promising solutions to remediation of other important pollutants such as arsenic or chromium.