Gene circuits to control, identify, and select cell populations
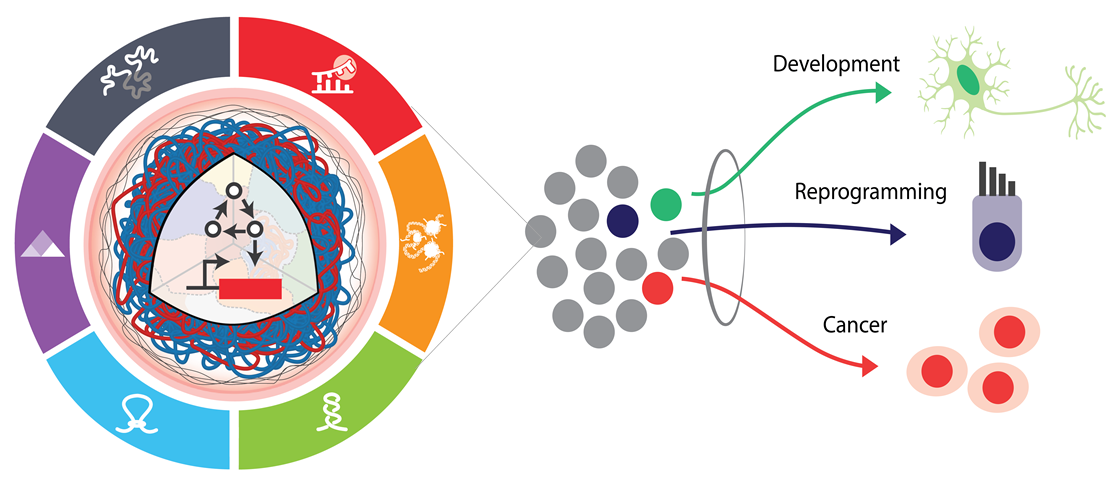
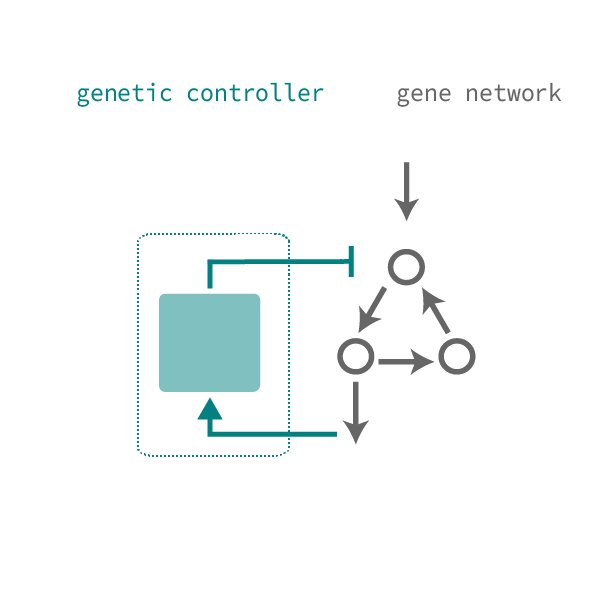
Objective: To interface with native networks to extract information on cell state and identity and to predictably actuate changes in transgene expression to identify, select, and control cell fates.
Why? Over the last decade, advances in genome engineering and stem cell biology have massively expanded the potential of engineered cells as therapeutics. To rapidly advance gene and cell-based therapies, synthetic biology aims to harness the power of native biology via gene circuits capable of dynamically prescribing cellular processes, states, and identities.
Challenges. Predictive performance and design as well as translatable tools and techniques remain limited for primary cells. Using primary cells as a testbed, we are developing novel tools for identifying, selecting, and controlling primary cells with gene circuits.
Vision. Using a combination of molecular engineering and computational modeling, we are improving the ability to predict circuit performance and to detect cell state and identity. These tools are accelerating the speed at which we can engineer cells and extract biological insights.
Papers
Model-guided design of microRNA-based gene circuits supports precise dosage of transgenic cargoes into diverse primary cells. Cell Systems (2025)
High-resolution profiling reveals coupled transcriptional and translational regulation of transgenes. Nucleic Acids Research (2025)
STRAIGHT-IN Dual: a platform for dual, single-copy integrations of DNA payloads and gene circuits into human induced pluripotent stem cells. bioRxiv (2024)
Supercoiling-mediated feedback rapidly couples and tunes transcription. Cell Reports. (2022)
Dynamically reshaping signaling networks to program cell fate via genetic controllers. Science (2013)
Reviews and perspectives
Bringing neural networks to life. Science. (2024)
RNA-based controllers for engineering gene and cell therapies. COIBT (2024)
The sound of silence: Transgene silencing in mammalian cell engineering. Cell Systems. (2022)
Synthetic gene circuits as tools for drug discovery. Trends in Biotechnology (2022)
Feedback loops in biological networks. Computational methods in synthetic biology. (2015)

