Molecular tools and models to engineer cell fate
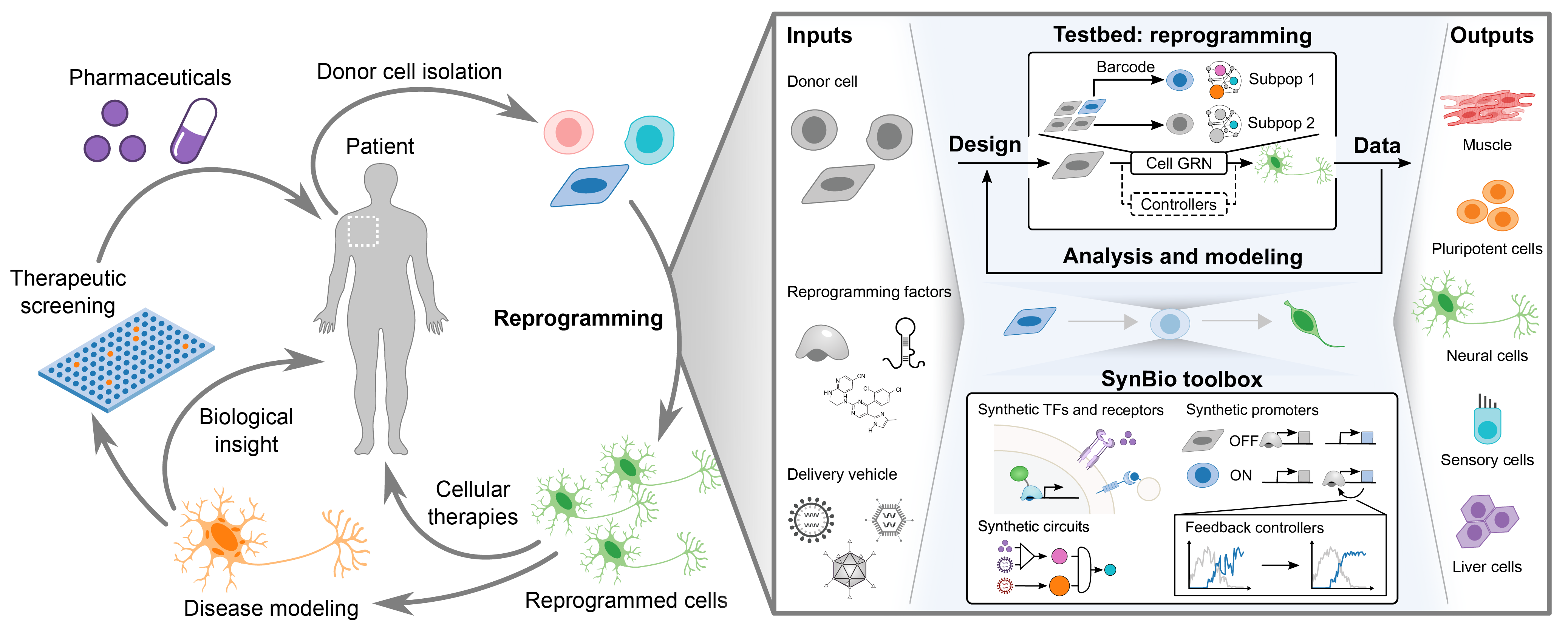
Objective: To develop molecular tools and models to understand and engineer high-efficiency cellular reprogramming using a model reprogramming system of fibroblast to motor neurons.
Why? Direct conversion via overexpression of lineage-specifying transcription factors confers synthetic genetic control over fate and can rapidly and efficiently induce neuronal identity.
Modeling cell fate transitions. Reprogramming via transcription factor overexpression provides a rich landscape for investigating the performance and potential of synthetic genetic systems to control cell fate in primary cells.
Challenges. While direct conversion provides a direct path to neurons and other inaccessible cell types, conversion yields have limited broad adoption. We developed improved reprogramming protocols and increased reprogramming rates for yields > 100%, improving the scalability and translational potential of induced motor neurons.
Vision. With improved protocols and minimized cassettes of reprogramming factors, we aim to examine the therapeutic potential of engineered neurons. Harnessing our novel synthetic biology tools, we are developing gene circuits to autonomously guide cells to new cell fates.
Papers
Compact transcription factor cassettes generate functional, engraftable neurons by direct conversion. Cell Systems. (2025)
Proliferation history and transcription factor levels drive direct conversion to motor neurons. Cell Systems. (2025)
Chemogenetic tuning reveals optimal MAPK signaling for cell-fate programming. bioRxiv. bioRxiv. (2024)
Cells transit through a quiescent-like state to convert to neurons at high rates. bioRxiv. (2024)
Mitigating antagonism between transcription & proliferation allows near-deterministic cellular reprogramming. CSC. (2019)
Comparative genomic analysis of embryonic, lineage-converted, and stem cell-derived motor neurons. Development. (2017)
Reviews and perspectives
Rewinding the Tape to Identify Intrinsic Determinants of Reprogramming Potential. (2024)
Accelerating diverse cell-based therapies through scalable design. Annual Review of Chemical and Biomolecular Engineering. (2024)
Changes in cell-cycle rate drive diverging cell fates. Nature Reviews Genetics. (2024)
Engineering cell fate: Applying synthetic biology to cellular reprogramming. COISB. (2020)

